Software
- We share most of our source code on https://github.com/CISMM.
- If you have any questions, please contact Joe Hsiao at phsiao@cs.unc.edu.
Software for microscope control
Fast 4D Volumetric Scan
mm_volumetric_scan.bsh (Micro-Manager 1.4)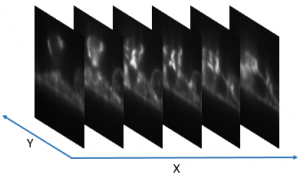
labview_volumetric_scan_continous.vi (LabVIEW 2016)
Two software tools are provided for fast 4D volumentric scans in Light sheet fluorescence microscopy. The first one creates a lookup table of light sheet position v.s. focus by performing an autofocus routine at each light position. The second one uses the lookup table and performs the high-speed volumetric scan. Each volumetric scan takes one second total, of two colors, which makes the videos of one frame per second in 4D.
AFM_sync_volulme_scan_MatLab.zip (including MatLab and LabView code)
We switched from Micro-Manager to MatLab, and this is the latest volume scan software.
SpinTIRF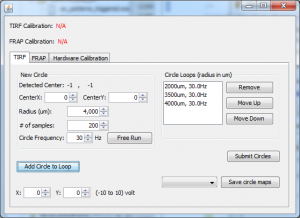
DualAxisMirror.jar ver 1.1 (64-bit Windows) (2/28/2017)
This plugin controls two-axis mirrors for applications like spinning TIRF or ring TIRF in Micro-Manager 1.4.
- Maps mirror coordinates to image coordinates.
- Allows users to define circles with arbitrary centers and radii and submit them to National Instrument boards.
- Synchronizes drawing circles in multiple channels with camera shutter if the NI board supports external triggers.
Note: To install just copy the .jar file to C:\Program Files\Micro-Manager-1.4\mmplugins
Optotune ETL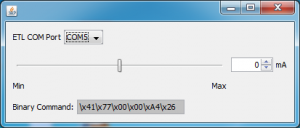
This is a Micro-Manager plugin that allows user to adjust voltages over Optoune electrically tunable lens to change its focal distance.
To use this plugin, one needs to add an “UserDefinedStateDevice” in MM for the ETL and
- Change the “Command Mode” setting to “Binary”.
- Change the “Port” setting to the port that ETL connects to.
Photoactivation
This is a script used in Micro-Manager for multiple-channel manipulation.
Script is also hosted on GitHub.
Active software
This section shows software that are still being used by us or collaborators.
Image Surfer (tutorials)
ImageSurfer 2.6.1-windows 64-bit
ImageSurfer 2.5.3 Mac 64bit (10.6.8 or newer)
ImageSurfer 2.5.3 Ubuntu 64bit
ImageSurfer 1.27 Windows 32bit
ImageSurfer is a software application used to view stacks of grayscale images, taken from a confocal microscope, as volumes. The user may choose different views–each offering a different interpretation of the data. In turn, the user may interact with the data as a three-dimensional volume or surface.
Video Spot Tracker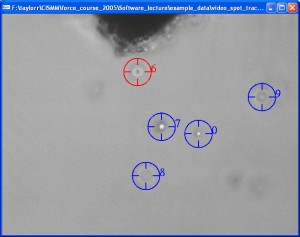
setup_video_spot_tracker_v08_01_02_extra_output
This version allows users to set a threshold on particle size to filter out undesired particles found in auto-find, e.g. aggregated beads. The area value associated with each particle is added in a new column in the output .csv file.
setup_video_spot_tracker_v08_11.exe (32-bit)
Bio-formats support has been added since v08.10.
Video_spot_tracker_v08_01a_MacOSX_Universal
setup_video_spot_tracker_v08_01.exe (32-bit)
The Video Spot Tracker program is used to track the motion of one or more spots in a set of image files, a video file, from a Microsoft DirectShow-compatible camera, or from some scientific cameras.
See the Video Spot Tracker manual for more information about the program. There are four tutorials available online:
Download the source code at GitHub.
Well Layout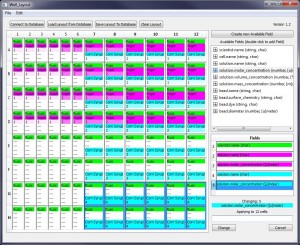
The Well Layout program lets you describe a 96-well experiment using a graphical user interface.
Layouts (and object in them) can be stored in a persistent Postgres SQL database or they can be stored in CSV-format files on a local hard drive.
Landmark Registration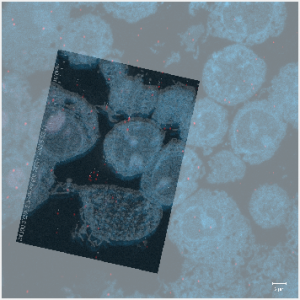
Landmark Registration for Windows 64bit
Landmark Registration for MacOSX 10.7 (Including Matlab Runtime)
This Matlab tool locates fiducials or markers in images from correlative microscopy and estimates transformations from them to achieve image registration.
The tutorial is here
Inactive Software
Software in this section do not have active users but we will still like to take feature requests.
Microscope Simulator
Microscope Simulator 2.3.0 for Windows XP/7 64-bit
Microscope Simulator 2.2.1 for Intel MacOS X 10.6
Microscope Simulator 2.2.1 for Windows XP/7 64-bit
Microscope Simulator 2.2.1 for Windows XP/7 32-bit
Microscope Simulator enables a scientist to simulate what a given 3D scene should look like under a fluorescence microscope. 3D scenes can be constructed from primitives or imported files in VTK, OBJ, or PLY format. Once a 3D model has been created, you can provide the point spread function (PSF) for a given fluorescence microscope and view simulated fluorescence images. Check out the tutorial.
The Microscope Simulator features an intuitive user interface and a multi-fluorophore optimization algorithm in the FluoroSim module.
Microscope Simulator 1.3.1 for Windows XP/7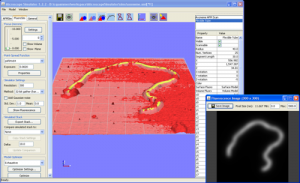
The Microscope Simulator enables a scientist to simulate what a given 3D scene should look like under a fluorescence microscope or an atomic force microscope (AFM). 3D scenes can be constructed from primitives or imported files in VTK, OBJ, PLY, or UNCA format. Once a 3D model has been created, you can provide the point spread function (PSF) for a given fluorescence microscope and view simulated fluorescence images. In addition, you can specify the parameters of a simple AFM tip model and quicky generate a simulated AFM surface scan.
See the Microscope Simulator web page for more instructions on using the program. A tutorial is available online.
ImageTracker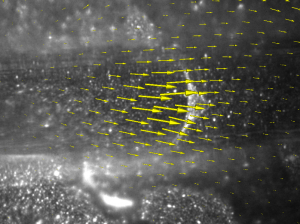
ImageTracker is a motion analysis application specifically designed for video microscopy. ImageTracker can stabilize image sequences with respect to moving objects, remove the effect of stationary components from bright field images, and compute and visualize dense displacement fields that explain motion in image sequences (optical flow). New in ImageTracker 3.0 is the ability to apply numerous image filters as well as an improved multi-resolution optical flow implementation.
See the ImageTracker web page for more information, including a manual. There are two tutorials available online: Image Stabilization and Removal of Stationary Image Components
Video Optimizer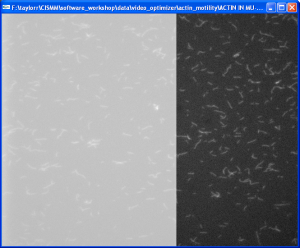
Video_Optimizer_2.03_IntelMac_Universal
setup_cismm_video_optimizer_02_03.exe
The CISMM Video Optimizer provides contrast and gain control, cropping, and point-based image stabilization on stacks of TIFF files and video files. It also enables simplified background subtraction and the accumulation of minimum, maximum, and average video frames.
Available online are a manual and two tutorials: Actin Motility and Mitotic-spindle Dynamics.
Template-Based Meatching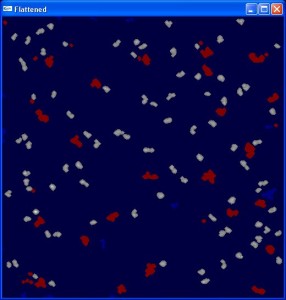
setup_template_based_matching_v01_05_1.exe
The CISMM Template-Based Matching program is designed to locate and identify objects within Scanned-Probe Microscope images and produce a file of descriptive statistics about these objects.
A manual and a tutorial are available online.
Network Extractor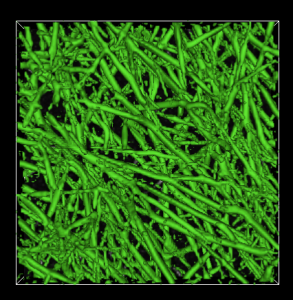
Network Extractor 1.0.1 for Windows XP/7 (64bit)
Network Extractor 1.0.0 for Intel MacOS X 10.5
Network Extractor 1.0.0 for Windows XP/7
Network Extractor is a program for segmenting and collecting statistics on fibrous objects in 3D images. It has been developed with the specific target of fibrin network images in mind, but can be applied to images of any kind of fibrous object such as collagen or actin networks, for example.
PSF Estimator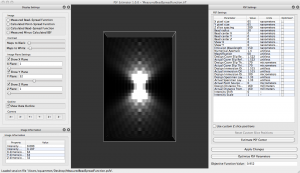
PSF Estimator 1.1.0 for Windows 7
PSF Estimator 1.1.0 for Intel MacOS X 10.6
PSF Estimator 1.0.0 for Windows XP/7
PSF Estimator 1.0.0 for Intel MacOS X 10.5
PSF Estimator is a program for fitting mathematical models of point-spread functions from microscopes to images of fluorescent beads with diameters below the resolution limit imposed by diffraction. Currently, only PSFs from widefield fluorescence microscopes are supported.
Source code is available here.
Simple Stereo
The Stereo Spin program is used to display stereo images from a spinning-loop video, making it easy to display stereo content from scientific visualizations or animations that were not originally rendered in stereo. It drives OpenGL stereo hardware if it is available. It can also drive cross-eyed stereo on any computer.
SketchBio
64-bit Windows installer for SketchBio version 0.0.10. See the installation guide for further instructions on how to set up SketchBio on your computer.
SketchBio aims to provide a rapid-to-use and easy-to-learn 3D modeling tool for biologists to enable effective interactive 3D “what if” scenario exploration for the exploration of subcellular structures. This is the source code for SketchBio.
Legacy Software
CISMM has a long history of creating software tools for the community. Software in this section haven’t been updated for a while but we find them are still helpful to users.
Plot Extractor
setup_CISMM_plot_extractor_02_05.exe
The Plot Extractor program is used to extract plot data from a TIFF image of a plot or series of plots. The data is saved in comma-separated values (.csv) format, which is readible by a number of programs including Excel and Matlab. The program currently runs only on Windows.
A manual and a tutorial are available online.
Edge Detector 1D
Edge Detector 1D is a collection of Matlab scripts that performs multi-scale edge detection on one-dimensional data. The download includes documentation on how to use the program.
A manual and a tutorial are available online.
Clarity
Clarity Deconvolution Library 1.0
Clarity is an open-source C/C++ library implementing many of the common deconvolution algorithms used in fluorescence microscopy. It is designed specifically for processing 3D images generated from optical sectioning. Clarity uses multithreaded algorithms to make full use of all the cores on modern multi-core computer systems. For even greater performance, the deconvolution algorithms can optionally run on commodity graphics processing units that feature hundreds of computing cores. Support for acceleration on graphics processing units is currently available on NVIDIA graphics cards.
A manual is available online.
The latest source code is available on GitHub.
WebSlinger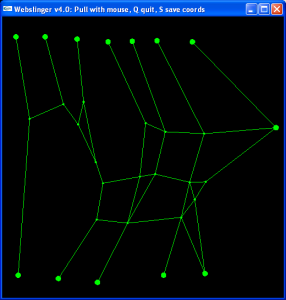
——————————————
webslinger-4.0.1 for Intel Mac
The WebSlinger program is used to simulate the deformation of a 2D web of masses connected by springs. The springs can be linear or nonlinear (with specified strain/force curves). The initial rest length and spring constant can be calculated based on the initial positions of the points and an initial strain estimate.
You can interact with the simulation by pulling on individual masses with the mouse. You can also set trajectories for one or more masses and compute the response of the other masses.
PreAnalysis
The PreAnalysis application will open a vrpn tracker log or a live vrpn tracker connection in order to calculate and display a mean-squared-displacement (MSD) plot.
Chromatin Cutter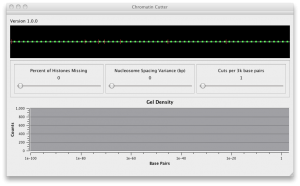
ChromatinCutter 1.0.0 Mac OS X Universal
This program simulates the distribution of DNA in a gel electrophoresis study of Yeast chromatin cut by enzymes. It lets you specify the fraction of nucleosomes that are unwrapped (wrapped ones have inaccessible DNA). It lets you specify the variance in the distribution of nucleosomes around their mean distance. It lets you specify the number of cuts per unit length (enzyme concentration). It plots the effects of these on the log number of base pairs, which matches the gel electrophoresis mass distribution. This provides a way to investigate whether features seen in the distribution are caused by nucleosome unwrapping or by nucleosome position variation.
Camera Calibration
Camera-calibration program brought to you jointly by CISMM.org and Megawatt Solar, Inc. This tool lets you print out a 2D color pattern that uses red and green to form a grid and blue to identify the grid, so that by selecting a few boundary points and thresholds you can automatically determine the camera’s intrinsic and extrinsic parameters. It uses Tsai’s coplanar method (source code included) to do the calculations once it has selected the points.
Spindle Simulation
Matlab source code and PDF manual for the Spindle Simulation published in the 2013 JCB paper “Pericentric Chromatin Loops Function as a Non-linear Spring in Mitotic Force Balance” with Andrew Stephens as the first author. This is a mechanical model of the behavior of the mitotic spindle of yeast during metaphase.
BrightPixel
BrightPixel is a plugin/script to Fiji (Fiji Is Just ImageJ) written in Jython that allows the user to track the movement of multiple fluorescent dots in 3D fluorescence micrograph videos. More abstractly, it tracks the motion of high intensity regions of pixels in a hyperstack opened in Fiji that defines a time series of z-stacked images.
nanoManipulator
nanoManipulator server for the Asylum microscope
This is the “External Operations” (XOP) file that provides a network interface for an Asylum scanned-probe microscope so that the nanoManipulator system can connect to it. This system was developed by 3rdTech when it sold the system through 2013. It is now released under the Boost open-source license. To use it, you also need to download the nanoManipulator installer. Contact support@nanomanipulator.org for the source code.
The nanoManipulator is a virtual-reality interface to scanned-probe microscopes. It was licensed to 3rdTech, who sold the system and support through 2013. It is now an open-source tool available free for use, licensed under the Boost software license. This is the installer for the application. Using it also requires installing the Asylum External Operations (XOP) file on an Asylum scanned-probe microscope, or building a network interface for another microscope. Contact support@nanomanipulator.org for the source code.
Video Averager
This installer includes a pair of video utility functions with desktop shortcuts. One of them computes an image that is the average of all frames in a video (or file sequence). The other computes the maximum pixel values across the video. It stores a composite image in the same directory as the files it is run on. You run the scripts by dragging the video file (or one of the images from a sequence) and dropping it onto the desktop shortcut.
VRPN Log to Matlab
vrpnLogToMatlabInstaller_05_00.exe
This utility converts VRPN log files from 3DFM-related programs into a format readable by MATLAB.
Mystery of the Sick Puppy
Installer for “The Mystery of the Sick Puppy”, an instructional module to enhance knowledge about viruses, their structure, and how they replicate.