Landmark/Fiducial based Registration Toolbox
Dept. of Computer Science, University of North Carolina at Chapel Hill
Tian Cao, Serdar Cakici, Marc Niethammer, Russell Taylor
1. Introduction: Correlative microscopy is an integration of different microscopy technologies including conventional light, confocal and electron transmission microscopy. The registration of correlative correlative microscopic images is quite challenging because it is a multi-modal, multi-scale and multi-dimensional registration problem. However, if there are fiducials or markers in the images from correlative microscopy and we successfully locate corresponding fiducials, we can estimate the transformation directly from them. In this project, we implement a toolbox for landmark based registration. We introduce this toolbox in the following sections.
2. Installation: The toolbox has two versions: standalone application and source code. This toolbox has been tested on Windows and Mac OS.
Standlone Application: You can run this version without matlab installed. But you need to install MATLAB Compiler Runtime library (MCR) following the instruction: https://www.mathworks.com/products/compiler/matlab-runtime.html.
You can download the application here.
Source Code: You need to add the common and output folders to matlab path, then run Registration.m. You can download the code here: Code.
3. Framework: The toolbox contains three parts: preprocessing, landmark/fiducial detection, landmark/fiducial registration.
Preprocessing: Because of the differences between the images we want to register, a preprocessing step is necessary. Preprocessing is a step aimed to simplify the landmark detection, to be specific, to roughly detect the landmark regions (a binary image). In this toolbox, the preprocessing methods are including extract different channels or grayscale of the image, binarize image with user defined threshold, crop image to remove text regions and inverse intensities to make the fiducials bright.
Landmark/fiducial detection: Form the preprocessing step, we can get the rough landmark regions. We use a simple yet effective method to detect the landmarks: we first calculate the connected components (CC) from the binary image, each CC corresponds to a landmark, then we choose the center of each CC as the location of the landmark. This method cannot get precise locations of the landmarks but the results is pretty good in practice [1]. We would like also to implement more complicated method for landmark detection [2].
Landmark/fiducial registration: For each detected landmark in both images, we first find its two nearest neighbors using euclidean distance to generate a triplet, and compute the ratio r of the distances of the two neighbors to the landmark. Then we can find the corresponding triplet in both images based on the ratio r. For each corresponding triplet, we can estimate the affine transformation matrix. We choose the best transformation model as the transformation result [1].
For more details of this algorithm, please refer to [1].
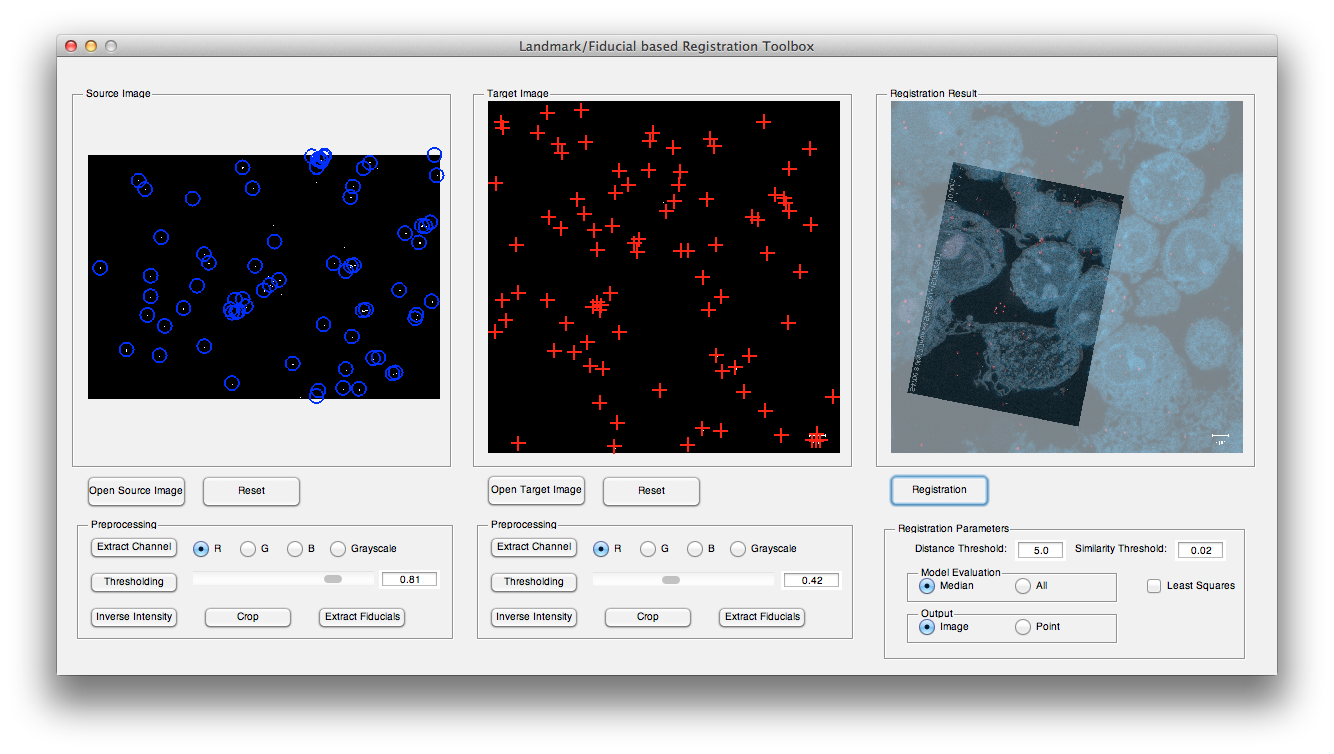
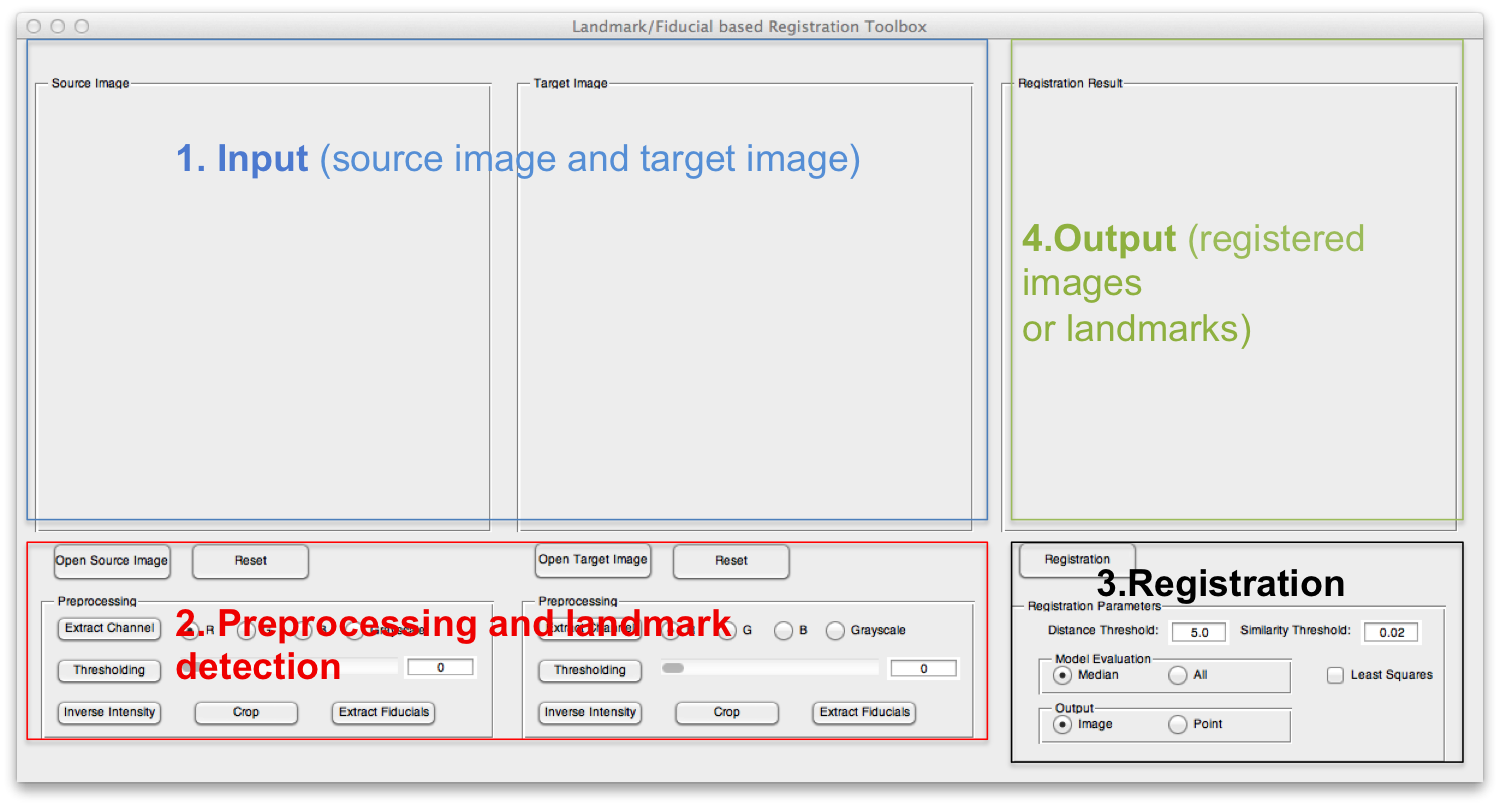
The GUI of the toolbox can be divided into four parts. Part 1 displays the source image and target image; part 2 can be used to load the input images and apply some necessary preprocessing methods and detect the landmarks; part 3 sets some registration parameters and part 4 shows the registration results.
A common workflow of landmark registration using this Toolbox is as follows,
a. Load input images: press Open Source Image and Open Target Image buttons to open corresponding source and target images. For example, open confocal_10.png as source image and macrophage_goldfiducial.png as target image from testdata folder.
b. Preprocessing: choose preprocessing method in part 2. For example, for source image, crop the image to remove the text on the lower boundary then apply a thresholding with 0.82, and for target image, extract the red channel and followed by a thresholding with 0.43.
c. Landmark detection: press both Extract Fiducials buttons to detect the fiducials in source and target images.
d. Registration: keep the default parameters then press Registration button.
5. Report bugs: please report bugs to tiancao@cs.unc.edu.
Enjoy the toolbox!
References:
[1] Tian Cao
“Multi-modal Image Registration for Correlative Microscopy,” PRP Report, Department of Computer Science, UNC Chapel Hill, 2011.
[2] Serdar Cakici.
“Marker-finding and automated registration in afm-fluorescence microscope hybrid,” PRP Report, Department of Computer Science, UNC Chapel Hill, 2010.