SketchBio
Overview:
Our collaborators often spend months or years developing complex geometric models and animation of subcellular complexes. We are making a tool, SketchBio, to reduce this to days.
SketchBio aims to provide a rapid-to-use and easy-to-learn 3D modeling tool for biologists to enable effective interactive 3D “what if” scenario exploration for the exploration of subcellular structures.
SketchBio development is being driven by collaborators with specific driving problems: the construction of protofibrils from fibrin monomers (Susan Lord) and the production of an animation of the mechanism of inhibition of focal-adhesion kinase (Vinay Swaminathan). We believe that developing a tool to specifically and completely solve multiple particular problems is the most rapid way to produce a generally-useful tool.
Immersive 3D Design
The primary goal is to make it easy for the scientist to produce a desired complex of 3D proteins and other structures, exploring how they fit together to form larger assemblies. This is intended both as a thinking aid and as a presentation aid.
Two-handed manipulation using a pair of six-degree-of-freedom magnetic trackers, provided by an inexpensive game controller. Collision detection between proteins and other structures, to enable precise placement without overlap. “Hollywood” physics including springs and potential fields to help objects maintain expected relationships. Links to external programs will enable realistic rendering of animations using Blender, coarse- and fine-grained simulations using SOFA or VMD.
By-Example Symmetry Transforms
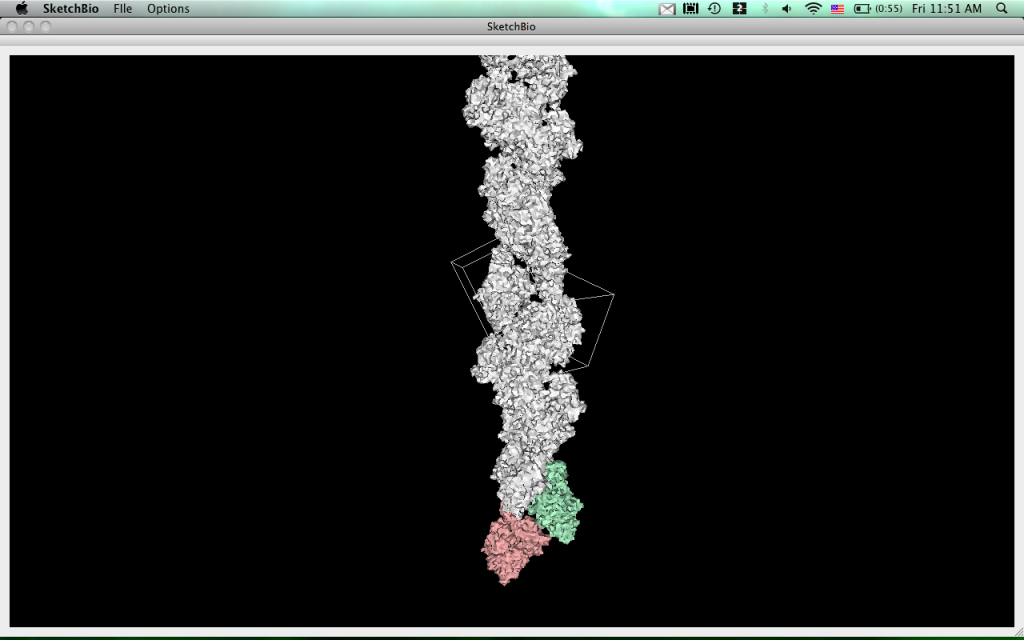
Many subcellular structures are composed of repeated copies of basic asymmetric units: fibrin protofibrils, microtubules, and virus capsids are examples of this. These structures can be constructed either by specifying the relationships among the copies by reading this information from a PDB file, by specifying the angles and offsets, or (often more naturally) by two-handed free-hand interaction with two copies of the molecule or by specifying desired proximities between the multiple copies. During free-hand and optimized placement, the entire crystal can be shown even while the manipulation occurs, to provide immediate feedback on the resulting structure. Figure 1 shows an example of an actin fiber constructed by repeating the transformation between the first and second monomers.
On-the-fly, Automatic Simplification
Structures with many proteins quickly become unwieldy to render and to compute collisions. SketchBio will use state-of-the-art simplification techniques to replace individual geometry edits with space warps, replace complex geometries with simplified ones, and replace long chains of molecules with tubular stand-in geometry during interaction. The resulting models and animations can be exported a full resolution for rendering or realistic simulation.
Status as of 10/21/2013:
The video shows some of SketchBio’s features that are helpful in making videos of macromolecular structures. These include collision detection, crystal-by-example, object replication, spring forces to bring molecules together at desired locations, animation keyframing, and fast-forward video preview.
Status as of 7/11/2013:
This video shows a user-defined animation where a set of actin monomers come together into a fiber, with a vinculin molecule approaching and touching the resulting strand. The animation was specified in Sketchbio using molecules loaded via Chimera and rendered in Blender — with all of the interaction and control happening from within Sketchbio.
Status as of 3/22/2013:
The current version of SketchBio can load arbitrary entries from the RSCB Protein Data Bank and fit them together in 3D. Many subcellular structures are composed of repeated copies of smaller asymmetric units. Examples of this are actin strands, microtubules, and fibrin protofibrils. We have implemented a method of copying the transformation from a pair of base objects to generate the rest of the structure (shown for an actin strand in image below). Spring-like bonds can be added between objects to constrain them to have certain relationships.
We are seeking additional collaborators with specific motivating problems who want to work with us as we develop the software. If you have a problem and are willing to help out on a development version of SketchBio, contact Russ Taylor or Shawn Waldon.
Programs used internally
To avoid rewriting existing code, SketchBio uses other programs internally to implement features whenever possible. For instance: interpreting a pdb file and generating a surface from it is done by running a script in UCSF Chimera. This surface can then be simplified (for faster rendering and collision detection times) using Blender, an open source modeling software.
Downloading
To install SketchBio on a Windows computer, download and the SketchBio Installer on our software page (read the installation guide for further instructions). The source code for SketchBio is available for download here, as well as on the software page.