Video Motion Extraction
The Challenge
Motion analysis is critical to understanding complex dynamic systems. Problems of interest to our research group include measuring the rate of mucus transport over lung epithelium cells, measuring the strain properties of the fibrin protein, and measuring the strain on a cell membrane induced by a moving magnetic bead.
Video microscopy presents unique challenges to traditional motion analysis techniques, including poorly-defined image features that appear and disappear, global intensity variations, low signal-to-noise ratios, and multiple layers of motion. Our goal is to provide a principled approach to formulating image analysis techniques that address these challenges.
The Approach
ImageTracker is an application written specifically for motion analysis in video microscopy. A Windows executable installer is available for download from the Center for Computer Integrated Systems for Microscopy and Manipulation (CISMM) web site here.
Image Filters
A chain of image filters modifies an image sequence loaded into ImageTracker before further analysis. Filters include intensity thresholding, Gaussian smoothing, gradient magnitude, and flat-fielding. ImageTracker uses the open-source Insight Toolkit (ITK) and Visualization Toolkit (VTK) to manage image loading, analysis, and display. This makes it easier to add more filters to future versions of ImageTracker.
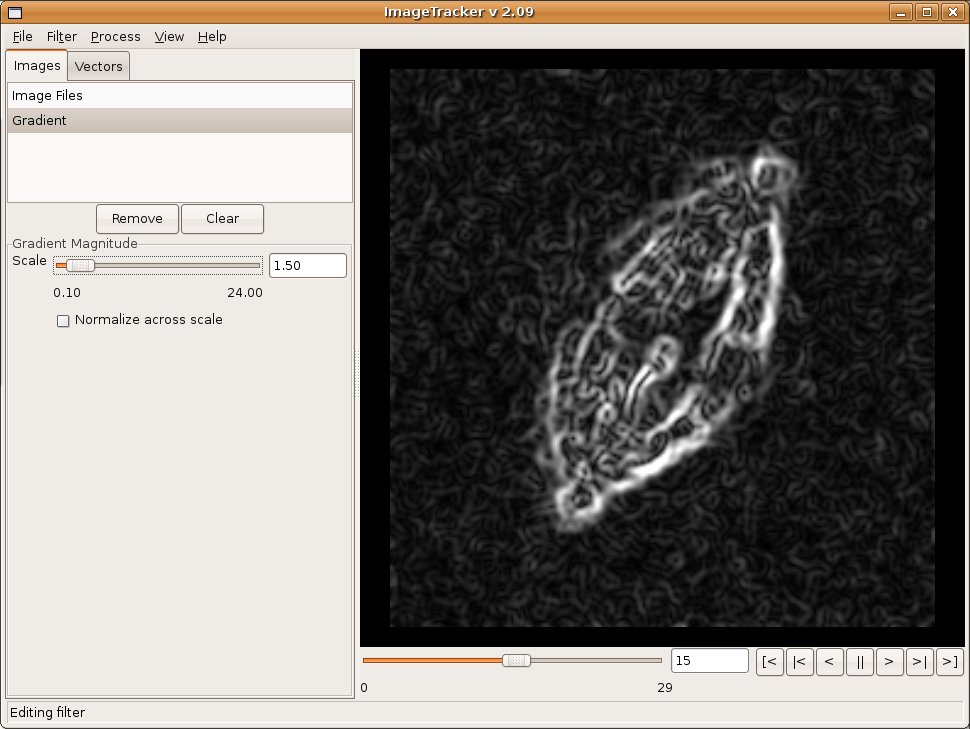
Image Alignment
Observations of in vitro specimens can suffer from the specimen moving across the field of view, which makes analysis of motion within the specimen difficult. ImageTracker includes multi-resolution image registration that stabilizes image sequences with respect to this global motion. The user selects the coarsest and finest level of detail to use for the image registration, which enables targeting specific types of motion. For example, selecting the coarsest scale to smooth the majority of image features aligns large image structures; selecting the finest scale to smooth small structures makes the alignment less sensitive to motion at that fine scale. After alignment, motion analysis of the fine structures can continue without concern for the global drift in the original video.
Background Removal
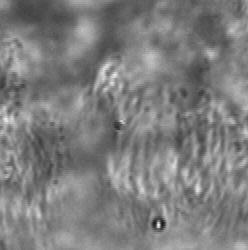
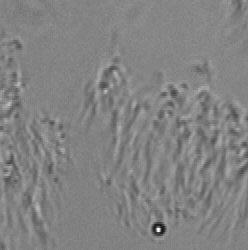
Long working distance lenses required for in vitro observation result in large depths of field, which means specimen elements outside the target observation plane contribute to the video. When focusing through a non-moving layer it can be difficult to interpret the motion in a moving layer. ImageTracker computes a model of the non-moving layer by compiling statistics on the gradient of transmitted light over the entire image sequence and integrating the estimated non-moving transmission gradient. This model enables computing the image that would have been formed with only the moving layer present. This method also corrects for non-uniform illumination and debris on the image sensor.
Optical Flow
Optical flow is a class of computer vision techniques for estimating the motion in an image sequence. Classic optical flow techniques use either local image calculations (which are more robust to noise) or global energy minimization (which creates a dense motion estimate). ImageTracker uses an implementation of an optical flow algorithm that combines local and global motion estimates (Bruhn et al., IJCV 2005). ImageTracker integrates frame-to-frame optical flow into net displacement fields.
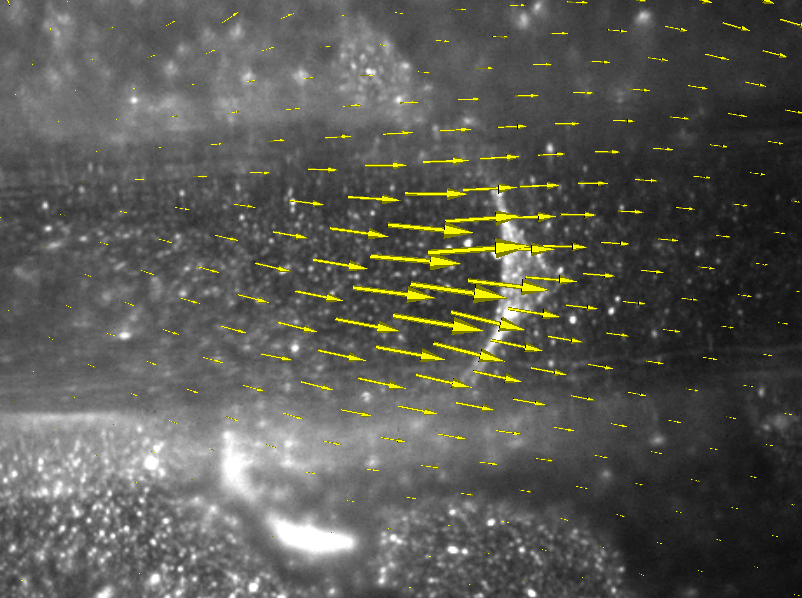
Flow Visualization
ImageTracker visualizes the computed optical flow field along with the original image sequence. Two visualization techniques are provided. Arrow glyphs represent flow magnitude (length) and direction at a sampling of image locations. Alternatively, a pseudocolored height map represents flow magnitude over the entire image plane.
Project Leaders
Russell M. Taylor II, research professor (Computer Science)
Richard Superfine, Bowman and Gordon Gray professor (Physics and Astronomy)
Other Investigators
Marc Niethammer, assistant professor (Computer Science)
Michael Falvo, research associate professor (Applied and Materials Sciences)
Tim O’Brien, research biologist (Physics and Astronomy)
David Hill, post doctoral researcher (Cystic Fibrosis Center)
Leonard McMillan, associate professor (Computer Science)
Graduate Research Associates
Brian Eastwood, Lamar Mair, Jeremy Cribb, Adam Shields, Vinay S. Swaminathan, Nathan Hudson, Daniel Millard (undergraduate)
Research Sponsors
NIH National Institute of Biomedical Imaging and Bioengineering (NIH P41-EB002025)
Selected Presentations of This Work
O’Brien, E.T., Falvo, M.R., Millard, D., Eastwood, B., Taylor, R.M., and Superfine, R. Monomolecular fibrin sheets: A possible kinetic intermediate in fiber assembly. Proceedings of the National Academy of Sciences USA, (to appear).
Eastwood, B. and Taylor, R.M. Occlusion removal in video microscopy. Computer Analysis of Images and Patterns, 2007, 4673, 125 – 132.
Eastwood, B. ImageTracker: Microscopy image analysis demonstrations. Light Microscopy for Biomedical Research Workshop, University of North Carolina, September 2008.
Eastwood, B. ImageTracker: Software for microscopy image analysis. Lasers and Microscopy User Conference, University of North Carolina, June 2008.
Taylor, R.M., Eastwood, B., Quammen, C., Feng, D., and Marshburn, D. Computer Integrated Systems for Microscopy and Manipulation (CISMM) Software Workshop. Force Measurement and Manipulation in Biological Microscopy, University of North Carolina, May 2008 and May 2007.
Keywords
Image analysis, computer vision, motion analysis, optical microscopy
For More Information
Dr. Russell M. Taylor II or Dr. Richard Superfine
Phone: (919) 962-1701 or (919) 962-1185
Fax: (919) 962-1799 or (919) 843-7308
Email: taylorr@cs.unc.edu or rsuper@physics.unc.edu