Registration for Correlative Microscopy
Correlative microscopy is concerned with the alignment of microscopy images from different modalities (such as the alignment of confocal with electron microscopy images) [1].
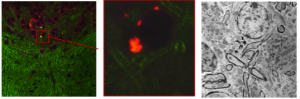
Our focus so far has been on learning how to transform (predict) the image of one modality into the other thereby transforming a multimodal into a monomodal registration problem. Our method is based on image analogies [2]. We use a sparse representation model to obtain image analogies. The method makes use of representative corresponding image training patches of two different imaging modalities to learn a dictionary capturing appearance relations.
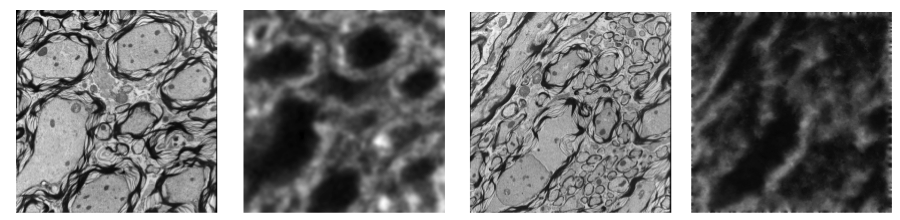
We also recently proposed a robust dictionary learning method for multimodal images. The crucial part in our image analogy method [2] is the learning of a joint dictionary for both modalities. However, dictionary learning may be impaired by lack of correspondence between image modalities in training data, for example due to areas of low quality in one of the modalities. Therefore, we proposed a probabilistic model that explicitly accounts for image areas that are poorly corresponding between the image modalities [3]. As a result, we can cast the problem of learning a dictionary in presence of problematic image patches as a likelihood maximization problem and solve it with a variant of the EM algorithm showing improvement in image prediction compared to the standard image analogy method.
References:
[1] J. Caplan, M. Niethammer, R. Taylor, and K. J. Czymmek, “The power of correlative microscopy; multi-modal, multi-scale, multi-dimensional,” Current Opinion in Structural Biology, 2011. [2] T. Cao, C. Zach, S. Modla, D. Powell, K. Czymmek, and M. Niethammer, “Registration for Correlative Microscopy using Image Analogies,” Workshop on Biomedical Image Registration (WBIR), 2012. [3] T. Cao, V. Jojic, S. Modla. D. Powell, K. Czymmek, M. Niethammer, “Robust Multimodal Dictionary Learning,” accepted to MICCAI 2013.