Mitotic Spindle under Brownian Motion
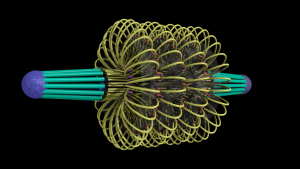
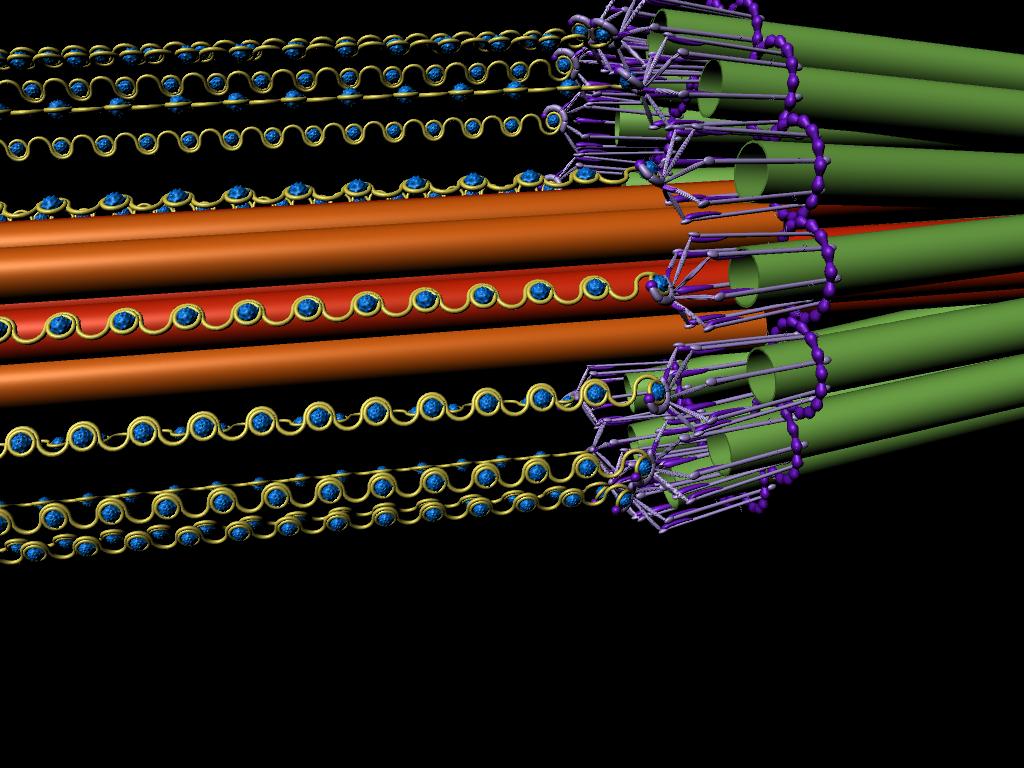
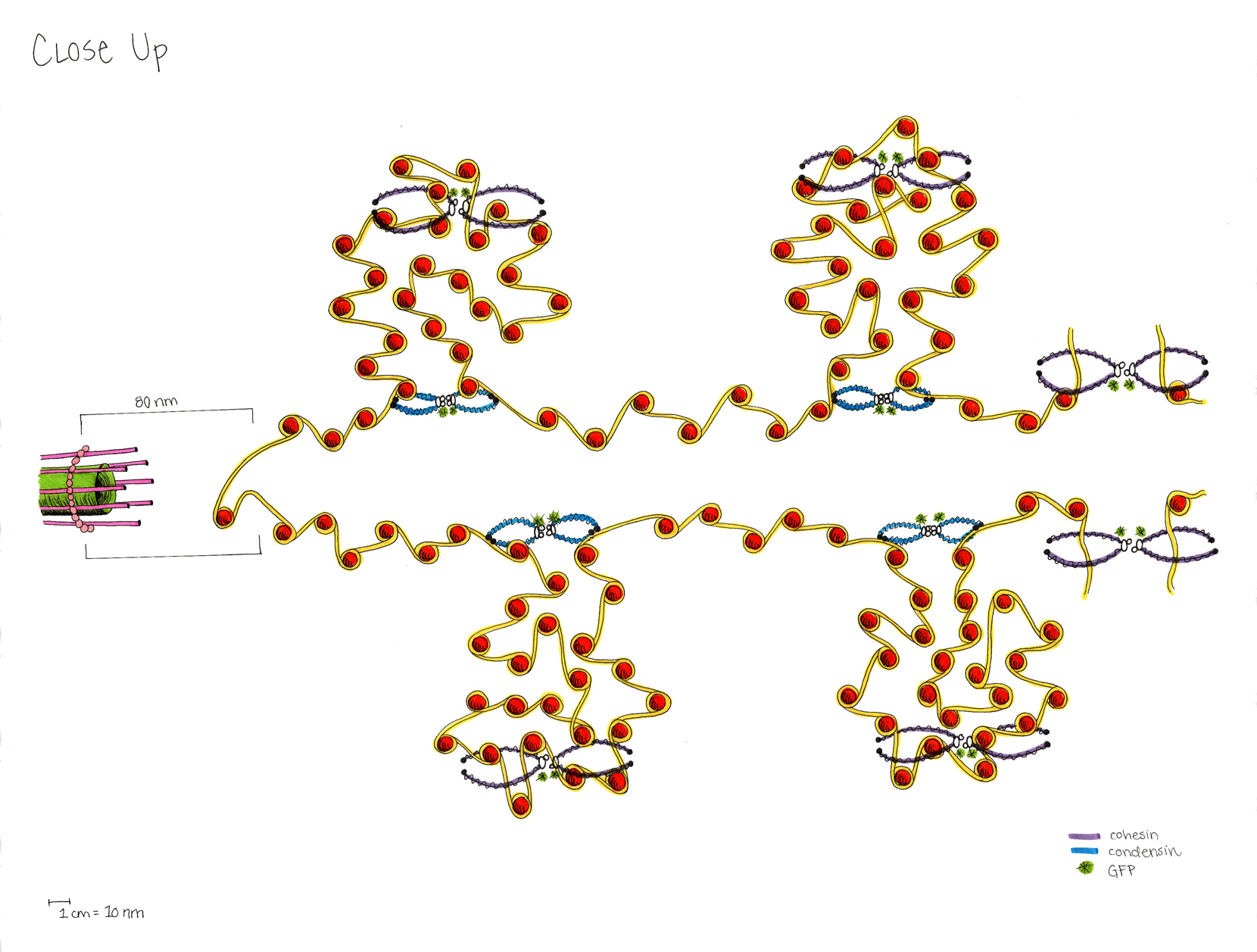
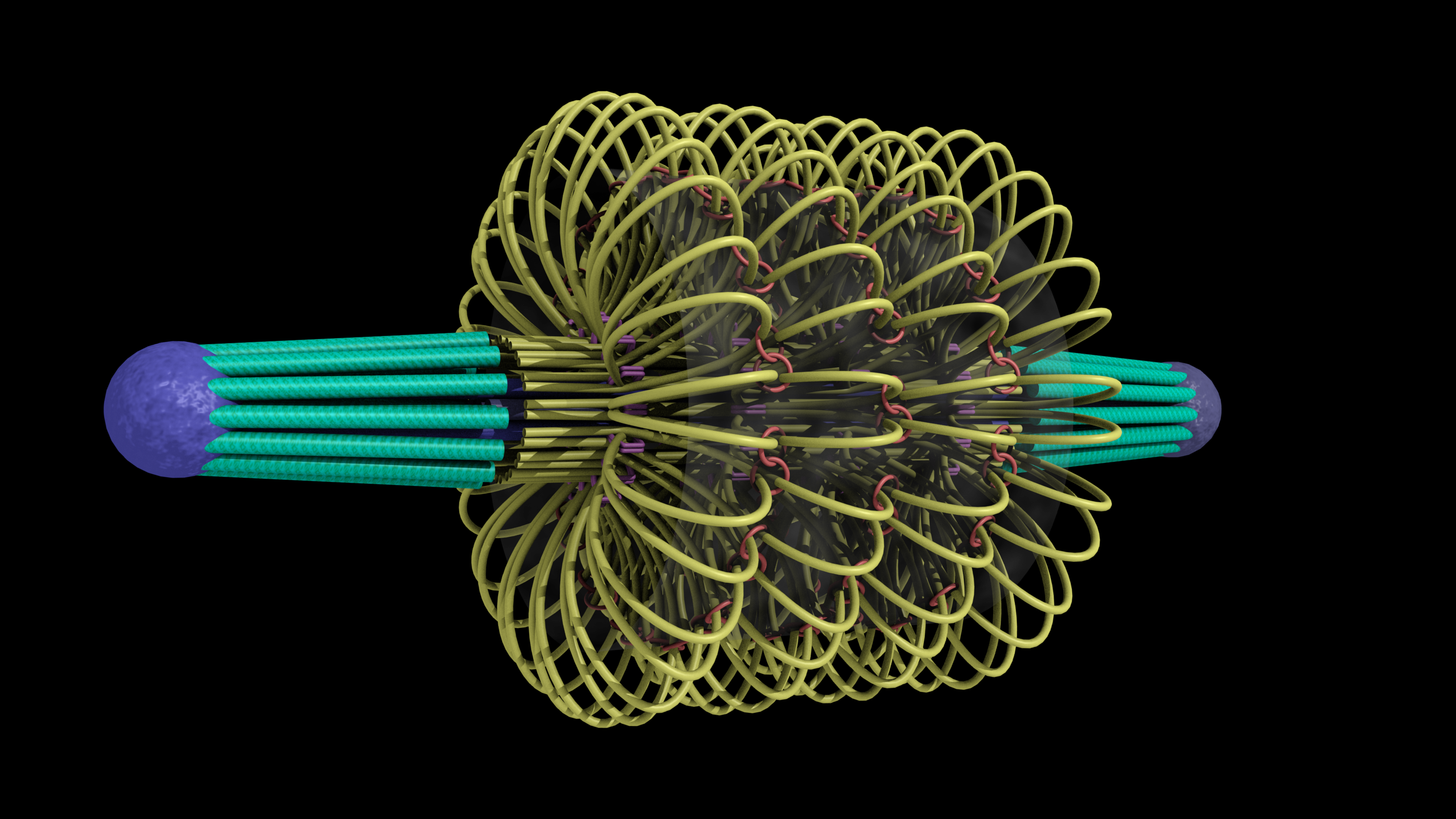
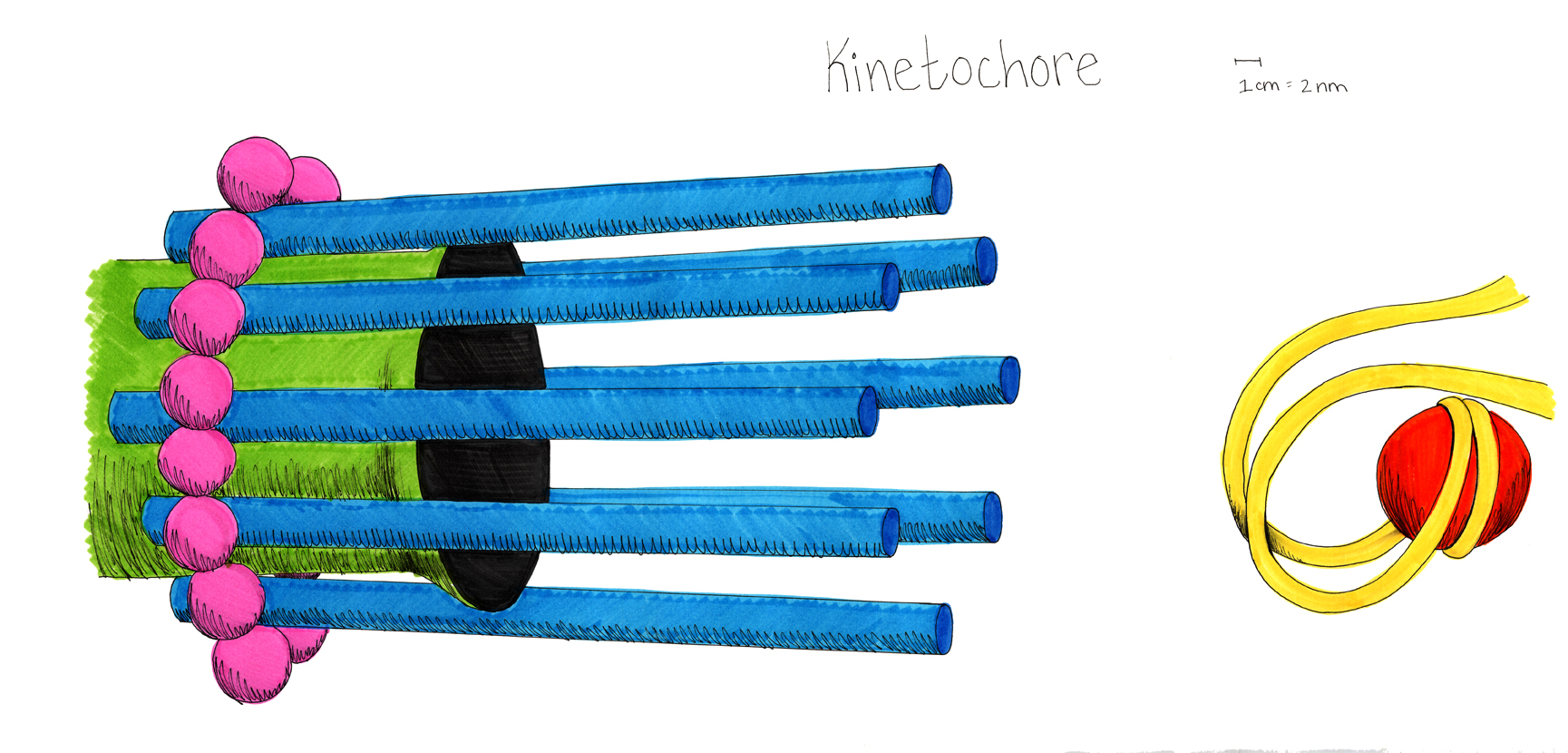
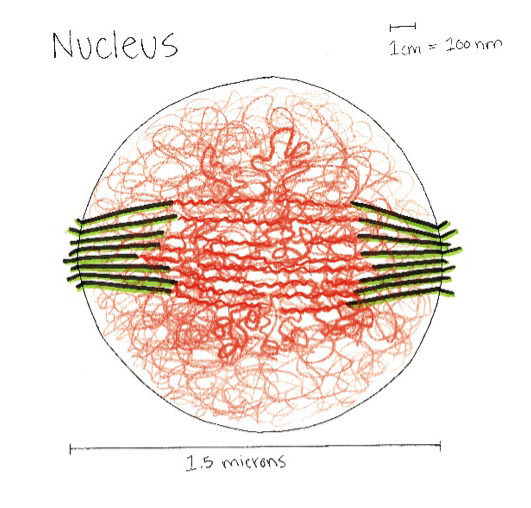
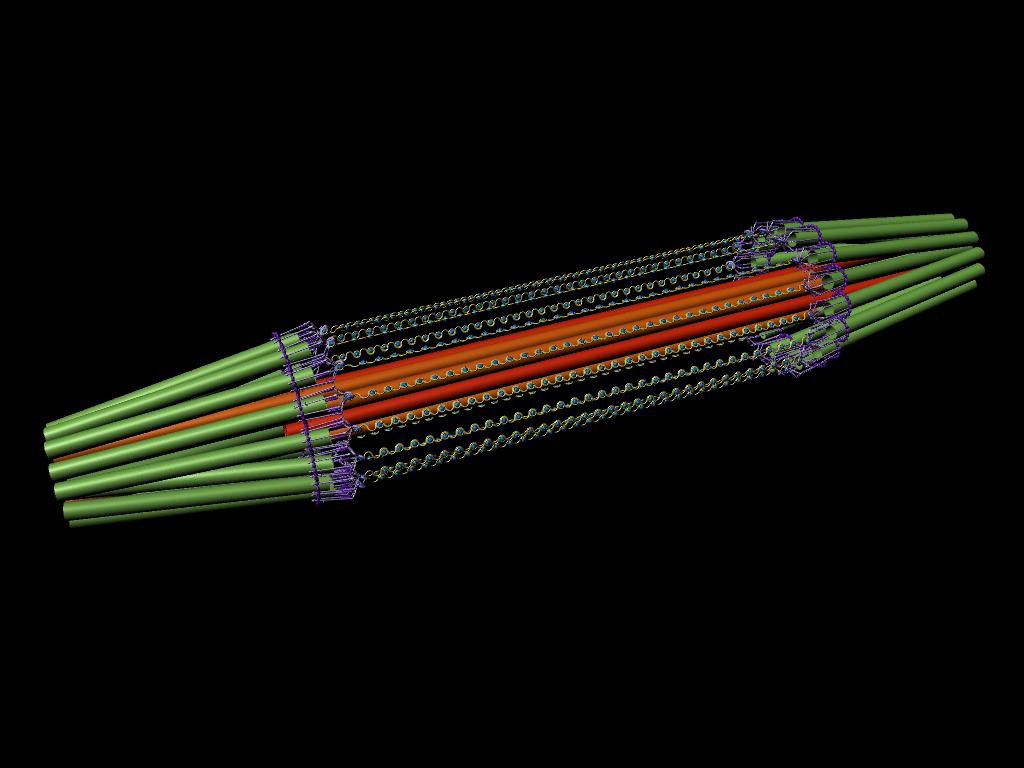
A model of the yeast mitotic spindle in metaphase was constructed based on a looping model of DNA distribution, where the loops are tied together at their base by condensin and they are linked by slip-rings of cohesin. This model was created in collaboration between Kerry Bloom and his group and CISMM personnel. The geometry of the model at its initial state is shown below.
The code was developed by Belinda Johnson. A coarse-grained simulation was run, based on the SOFA platform with the addition of Brownian forces and stiffness forces, to understand what the addition of Brownian forces on the structure would do to the distribution of DNA. The parameters of the simulation included a random force of 1228, a mass_damping of 10, a mass_radius of 0.0045, a spring constant of 628000, a hinge force of 0.548, and an object mass of 33.3 (see simulation code for the units). We believe that this model matches the expected behavior except for: (1) the viscosity is much lower than in reality so that the simulation will run in reasonable time (we expect the behavior to be the same). (2) The cohesin rings are too large by about a factor of two (this was a mistake in the model set up, which we think will only serve to make the rings more mobile). (3) There is no torsional restoring force on the DNA, so there will be no impact due to twisting of the DNA strands.
The resulting movie of the simulation is available here: 0001-0167. It shows a snapshot of the simulation every 0.1 nanoseconds up to 17.6 nanoseconds.
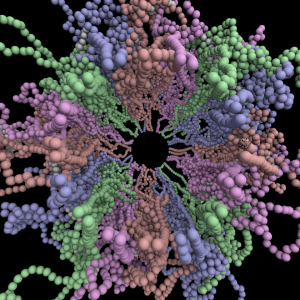
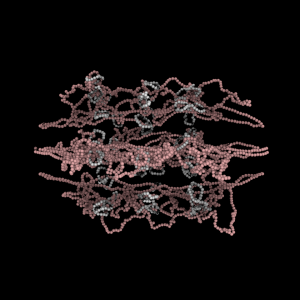
The simulation showed the outside of the rings pulling in and the inside of the spindle pushing out. There was not a large net offset to the cohesin rings from their initial position (each moves, but they seem to maintain their approximate position). The ends of the DNA are pinned in space as if to stable microtubules. Some images of the resulting configuration are shown below.