Yeast Mitotic Spindle
The image below is a proposed structure for the mitotic spindle of yeast during metaphase that was produced in a collaboration between Russell M. Taylor II, Andrew Stephens, Kerry Bloom, Leandra Vicci, Jolien Verdaasdonk, Steven Nedrud, Matt Larson, and Michael Falvo.
Others participated in the earlier development of the model, including Kendall McKenzie and Callie Holderman.
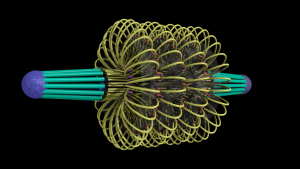
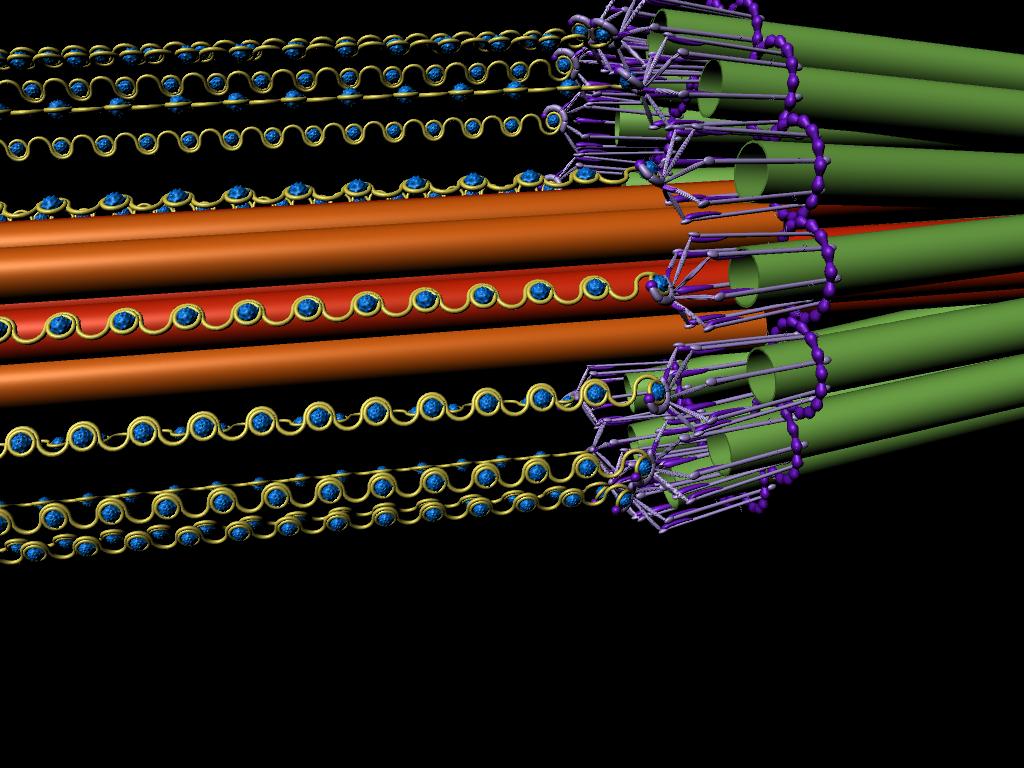
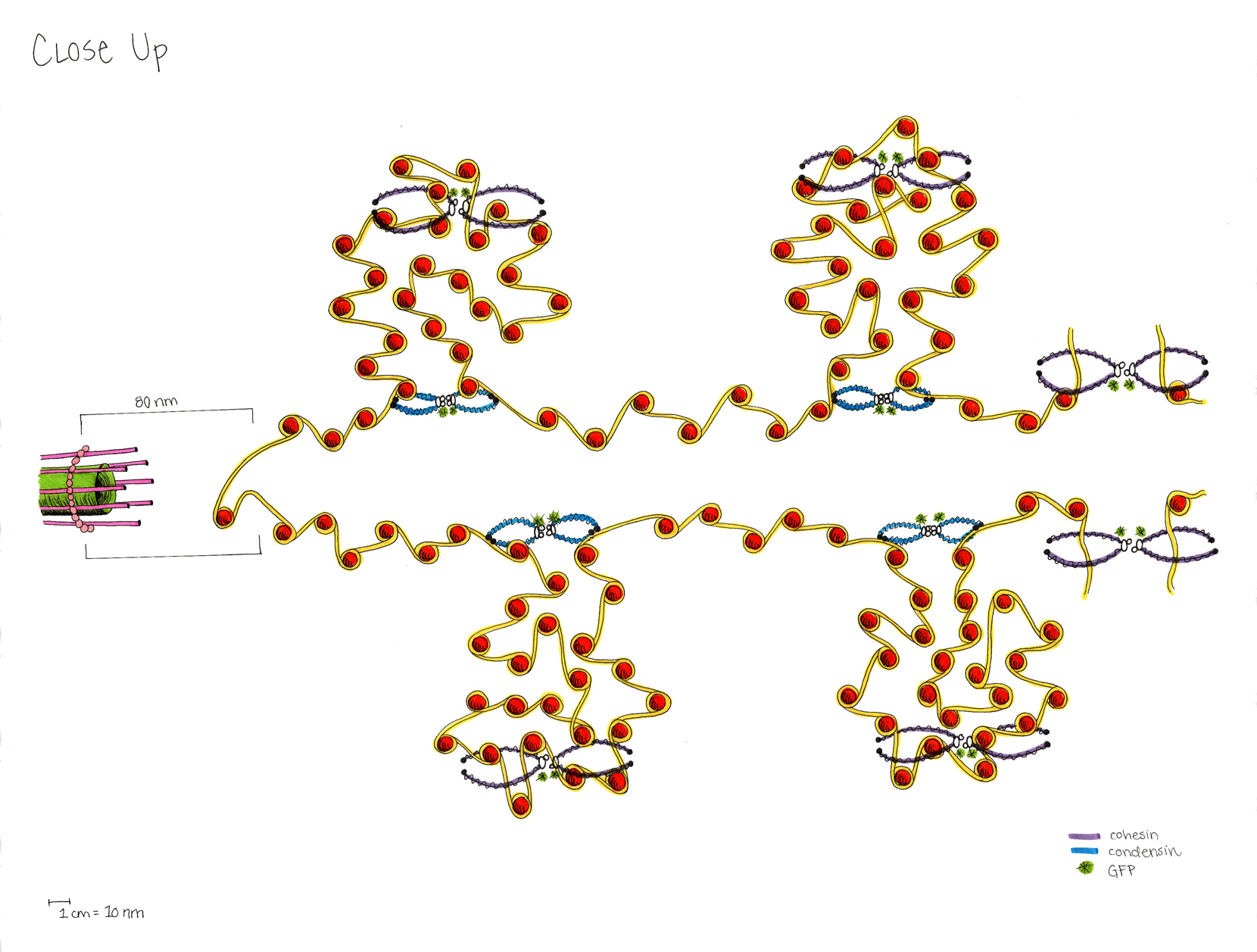
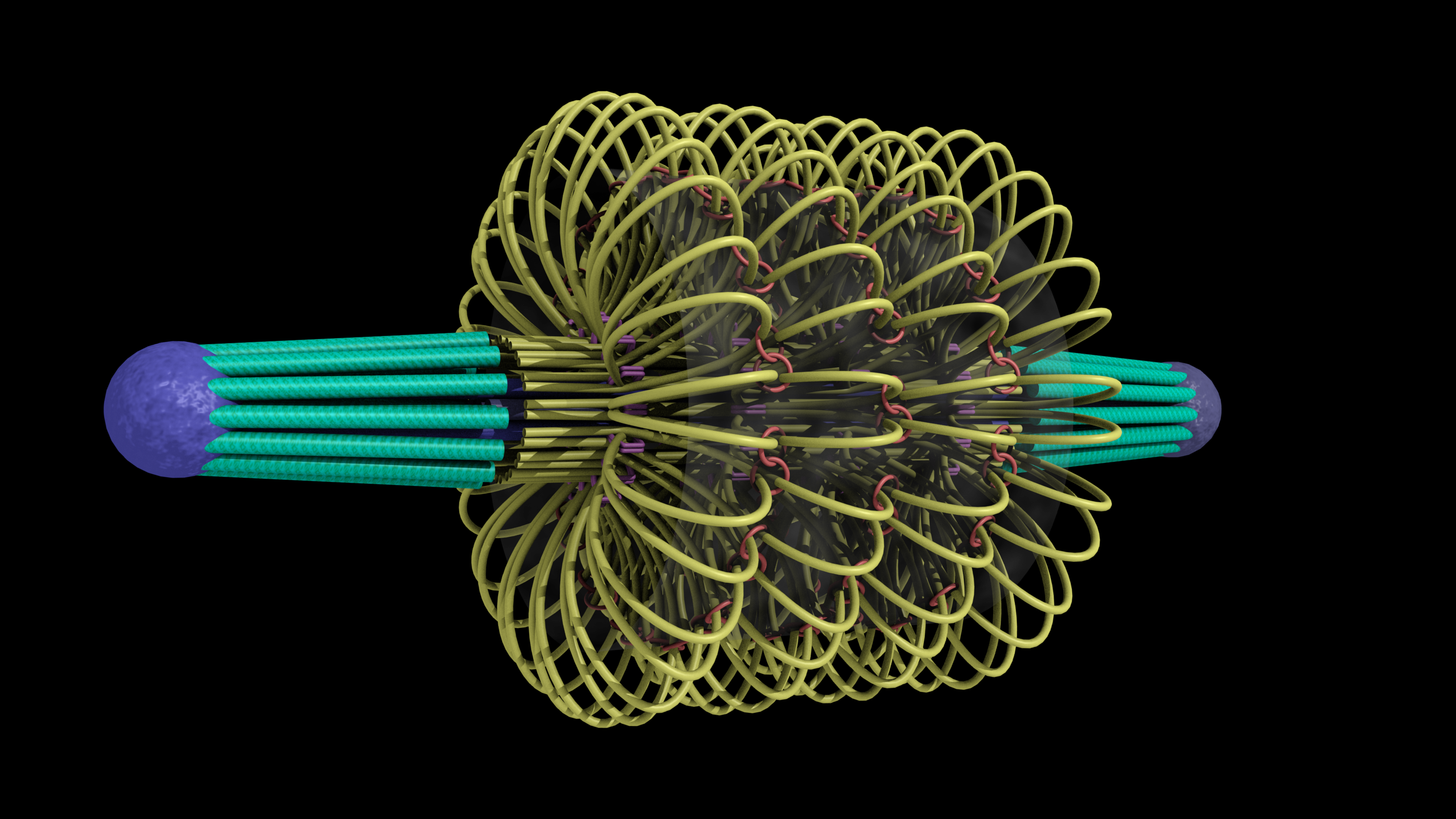
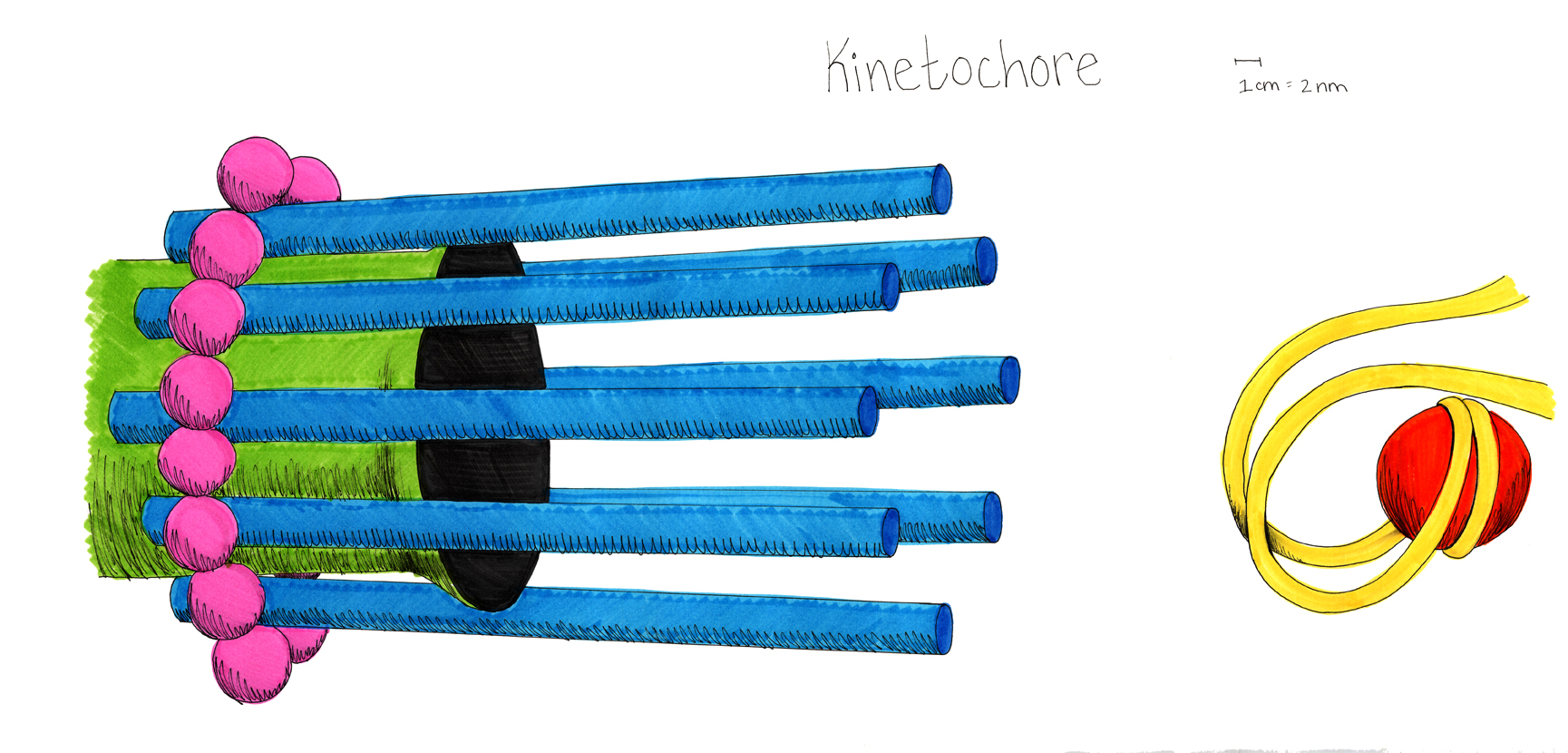
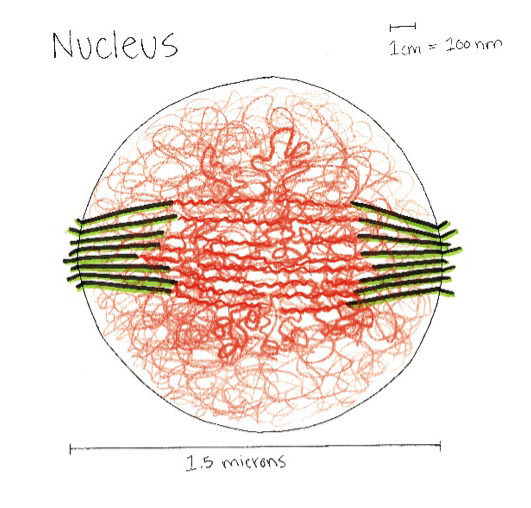
This model of the yeast mitoric spindle shows the spindle-pole bodies as blue spheres, the kinetochore microtubules as green cylinders, the DNA as yellow tubes, cohesin as linked red rings, and condensin linking molecules in purple. The translucent gray shell around the spindle shows the center of the region that contains cohesin as seen in fluorescence microscopy images taken of the spindle. The DNA and other structures are too small to be resolved in the microscope. This is the twentieth version of the model, which has been developed over a two-year period of intense collaboration between cell biologists, computer scientists, physicists, and artists. This was developed as part of the Computer-Integrates Systems for Microsopy and Manipulation NIH/NIBIB National Research Resource.
It is a geometric model of a hyopthetical structure that has evolved to be consistent with a number of experiments performed in the department of Biology. Its purpose is to display, in a consistent 3D space, a model, all known aspects of the structure. This forms a basis for discussion, which then results in new planned experiments and in changes to the model.
This image was an honorable mention in the illustration category of the NSF/Science visualization challenge 2010.
About the yeast: The yeast is Saccharomyces cerevisiae as used for baking and brewing. The strains we use are science based versions not commonly used for brewing or baking, but they are the same species. S cerevisiae is one of the most commonly used eukaryotic model systems in biology.