Migrating soon to website for Superfine Lab
Much of this content will soon migrate to a campus-hosted website for the Superfine Lab. Stay tuned!
Much of this content will soon migrate to a campus-hosted website for the Superfine Lab. Stay tuned!
We have developed a new imaging system that enables real-time X-Z imaging simultaneous with traditional X-Y imaging. This is done by arranging a 90º reflecting optic above the specimen within the field of view of the standard epi-fluorescence imaging system. We call this method PRISM, for Pathway Rotated Imaging for Sideways Microscopy. We use a freely positioned reflecting optic that can observe a wide variety of specimens, with standard imaging microscopes, integrating it with light sheet microscopy and atomic force microscopy.
This movie shows a side-view of the nucleus of a SKOV cell, an ovarian cancer cell line, with the DNA labeled with a red fluorescent nucleic acid stain, as captured with Vertical Light Sheet Sideways Microscopy (VLSSM). Over the course of the movie an Atomic Force Microscope (AFM) cantilever with 5μm bead tip deforms the nucleus with 50nN of force. The movie was recorded at 40fps with simultaneous force data acquired by an AFM. For more information on VLSSM follow this link : http://cismm.web.unc.edu/core-projects/force-microscopy/scanning_probe_technologies/
The video below shows some of SketchBio’s features that are helpful in making videos of macromolecular structures. These include collision detection, crystal-by-example, object replication, spring forces to bring molecules together at desired locations, animation keyframing, and fast-forward video preview. More information on SketchBio is available at sketchbio.org.
Kerry Bloom’s lab is collaborating with CISMM to understand the structure and forces in the mitotic spindle of yeast.
A geometric model of the mitotic spindle of yeast is challenged by the addition of physically-based forces. The resulting simulation reveals a complex mixture of symmetry and chaos. Color, motion, and careful removal of substructures are used to reveal fundamental behaviors. The model is up to the challenge — the location of ring-like cohesin molecules remains steady against the pounding of external molecules and the stiffness of the DNA. Pressure and entropy counteract one another in a careful balance. A multidisciplinary team of biologists, physicists, computer scientists, applied mathematicians, and engineers worked together to develop and refine the presented model. A supercomputer was used to simulate the motion, and then again to enable rendering of the complex, changing, translucent video sequence. Plain-language narration describes the behavior, while the simulation is repeated from various viewpoints and with varying rendering styles to provide clear explanation.
The 147MB 1280×960 Mpeg2 video file can be found here: Yeast Mitotic Spindle Model
CISMM has the honor of having won Honorable Mention in the NSF/Science Visualization Challenge three times. Michael Stadermann submitted an AFM image of a buckled carbon nanotube made with our nanoManipulator system in 2003. Russell Taylor, Briana K. Whitaker and Briana L. Carsten submitted an SEM image of our flexible post technology in 2009. The Mitotic Spindle Group submitted a computer model of the yeast mitotic spindle in 2010. Click on each image below for a higher-resolution version.
This video shows a computer rendering of the Panoptes system, made by Christian Stith using the CAD models from Leandra Vicci. It starts with the entire system, then shows how the light moves through the varioptic lens element and causes fluorescence in a single channel. It then shows the second fluorescence channel. It then reassembles the entire system and shows how it works, translating to each new position and exposing the two fluorescence channels to grab two images for each channel.
A model of the yeast mitotic spindle in metaphase was constructed based on a looping model of DNA distribution, where the loops are tied together at their base by condensin and they are linked by slip-rings of cohesin. This model was created in collaboration between Kerry Bloom and his group and CISMM personnel. The geometry of the model at its initial state is shown below.
The code was developed by Belinda Johnson. A coarse-grained simulation was run, based on the SOFA platform with the addition of Brownian forces and stiffness forces, to understand what the addition of Brownian forces on the structure would do to the distribution of DNA. The parameters of the simulation included a random force of 1228, a mass_damping of 10, a mass_radius of 0.0045, a spring constant of 628000, a hinge force of 0.548, and an object mass of 33.3 (see simulation code for the units). We believe that this model matches the expected behavior except for: (1) the viscosity is much lower than in reality so that the simulation will run in reasonable time (we expect the behavior to be the same). (2) The cohesin rings are too large by about a factor of two (this was a mistake in the model set up, which we think will only serve to make the rings more mobile). (3) There is no torsional restoring force on the DNA, so there will be no impact due to twisting of the DNA strands.
The resulting movie of the simulation is available here: 0001-0167. It shows a snapshot of the simulation every 0.1 nanoseconds up to 17.6 nanoseconds.
The simulation showed the outside of the rings pulling in and the inside of the spindle pushing out. There was not a large net offset to the cohesin rings from their initial position (each moves, but they seem to maintain their approximate position). The ends of the DNA are pinned in space as if to stable microtubules. Some images of the resulting configuration are shown below.
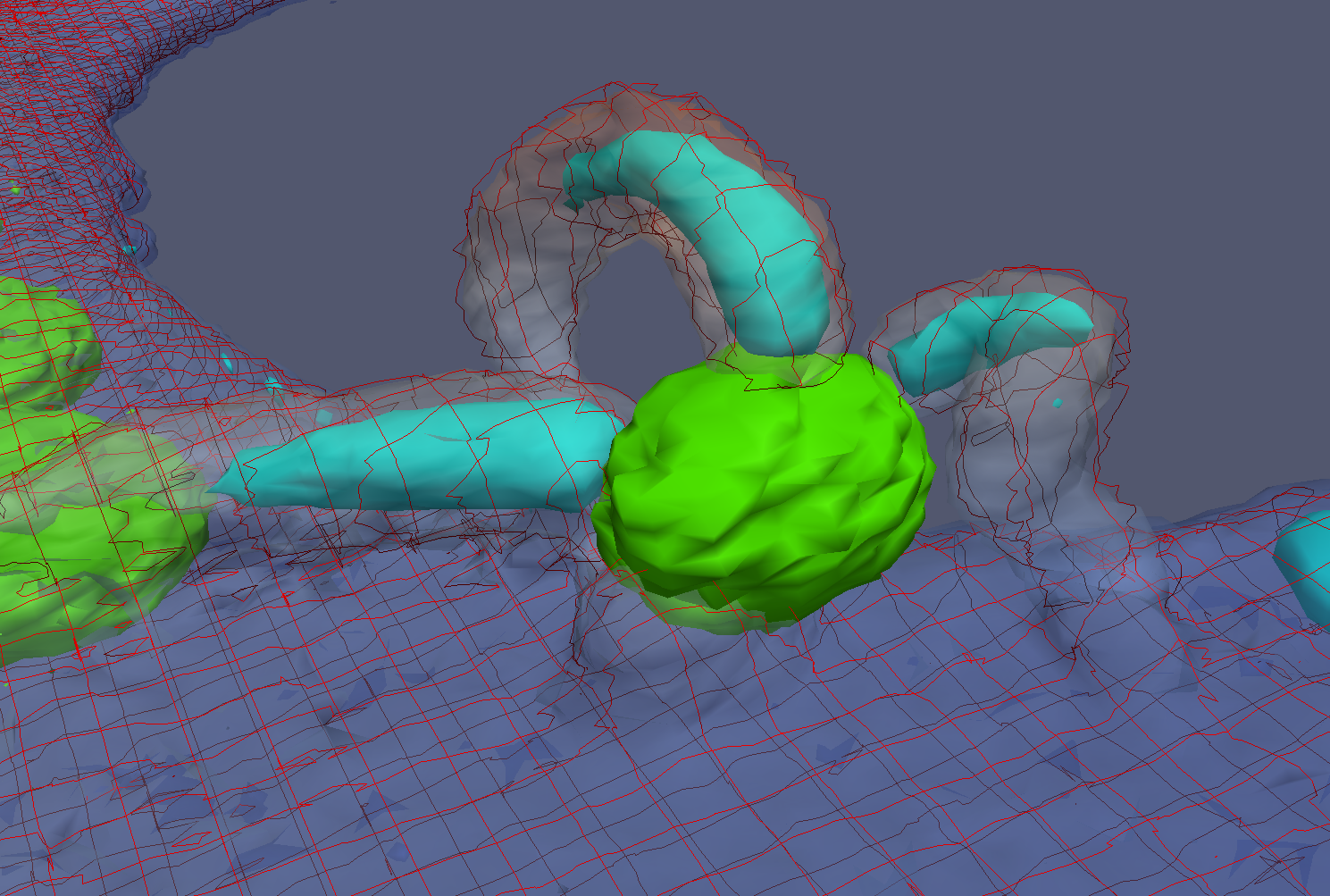
Collaborator Aaron Neumann from the University of New Mexico is studying fungipods from human dendritic cells that attach to yeast. The image above shows a combination of three different fluorophores that together show the behavior. A green yeast is sitting on top of the cell membrane (transparent red) with three fungipods attached to it. The fungipods are very dense in actin (blue).