Video Optimizer Tutorial: Spindle Dynamics
Data set and source
This data set of spindle dynamics is provided by Lisa Cameron at Ted Salmon’s laboratory at UNC Chapel Hill. It was provided to the CISMM Resource as an example of a mitosis-like event that has whole-spindle motion in addition to microtubule motion and they have agreed to let us distribute it as a data set to be optimized.
This data set is a series of time points of a mitotic spindle formed in a cellular extract from frog eggs. Three frames from the unoptimized video are shown here:



Suggested procedure for optimizing
Turn on gain control and set it to auto-gain. Set “reorient” and then put two cone trackers tracking white spots (turn off dark_spot toggle) on the ends of the spindle and set the radius of the trackers each to 30 pixels.
Enable optimize.
Check the show_gain_control checkbox, which will bring up a gain control panel; on that panel, enable the auto checkbox.
Turn off 16-bit logging. Set the base name of the output files. Press “play_video” to begin optimization. Three frames from the resulting optimized video are shown here:
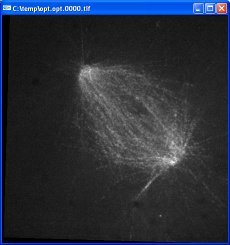
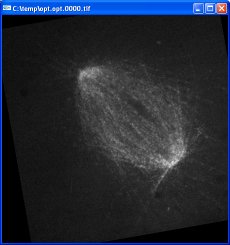
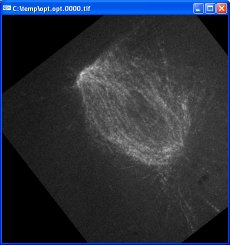
The black slanted borders in the optimized images are caused by regions where there is no data; these areas were outside the original images once they have been translated and rotated. The contrast in the optimized images is greater because of the auto-gain feature. The position of the upper-left spindle end remains fixed due to the optimization of the first tracker. The orientation of the spindle remains similar because of the reorient checkbox.