Creating fibrinogen chains in Chimera
This tutorials teaches you how to create chains of fibrinogens in the above image in Chimera. Here is its rotation movie.
Pull out the data
Click on the menu, Tools->General Controls->Command Line
Eexecute the following commands in order to download objects from pdb.org.
open 3ghg open 3ghg open 3ghg open 3h32 del :.G-L,.Q-T
3ghg is the fibrin in the chain, and 3h32 is a dimmer that we will use for reference when doing alignment.
Click menu, Tools->High-Order Structure->Multiscale Models, click button “Make models” at the lower left.
Close Mutiscale Models dialog.
Notice that the ribbons will now become surfaces. Actually, the ribbons are still there, but hidden for now. Click Actions->Ribbon->show to make they show up.
We change the representation to surfaces so we can see better during alignment. However, the ribbons are what we need in computing the matrcies later. It is important to move both the ribbon and surface when you move a fibrin.
Go to the bottom of main window, check 1 and 5 and uncheck the rest in “Active models”. Put the mouse cursor over the fibrin surface and use the middle button to drag surface #5 and ribbon #1 out. Do it again with only #2 and #6 checked this time to drag #2 and #6 out. Your screen should look like the image below. I labeled each fibrin the associated ribbon and surface numbers.
Initial alignment
Select #1 and #5 as Active models again, and move and rotate them next to surface #4. You may look at the dimer for reference.
Select #2 and #6, rotate and move them to the place you like.
ATTENTION:
Don’t change the setting in Tools->Movement->Movement Mouse Mode. If you use the setting of “Move molecule” to move surfaces, the actual transformation matrix is not going to be modified, even though it looks like the surfaces are moved on screen. We need the matrix to be modified to replicate more surfaces later.
The image below shows my initial alignment.
Duplication
Click Tools->Utilities->Reply Log to open a log window.
Back to the command line, type
measure rotation #0 #2
The command will display the matrix information of #2 relative to #0 ribbon in the Reply log window we just opened. My log is:
We got buch of information here, which is what we need in the sym command.
The format of sym command is “sym <obj to copy> group h,<shift along axis>,<Rotation angle>,<number of duplications (including the original)> axis <Axis> center <Axis point> coord <reference obj>”
By copying the corresponding fields, my sym command is like:
sym #2 group h,-213.41756905,110.80567673,19 axis -0.94964696,0.08108266,0.30264872 center 0,-21.81924641,-65.97804975 coord #1
After the sym command, I will have 18 more duplications of fibrins on my screen, with the same transformation of surface #2 relative to #0, starting from ribbon #1.
Next we want to delete surface #5 (ribbon #1) and the dimer, cause we don’t need them anymore. Hold Ctrl+shift, and use mouse left click to select surface #5 and the dimmer like in the image below. Tips: First select parts of both objects and then press the arrow key “up” on the keyboard to extend the selection to the whole objects.
Click Actions->Atoms/Bonds->delete to delete those objects. The final fibrins will look like this:
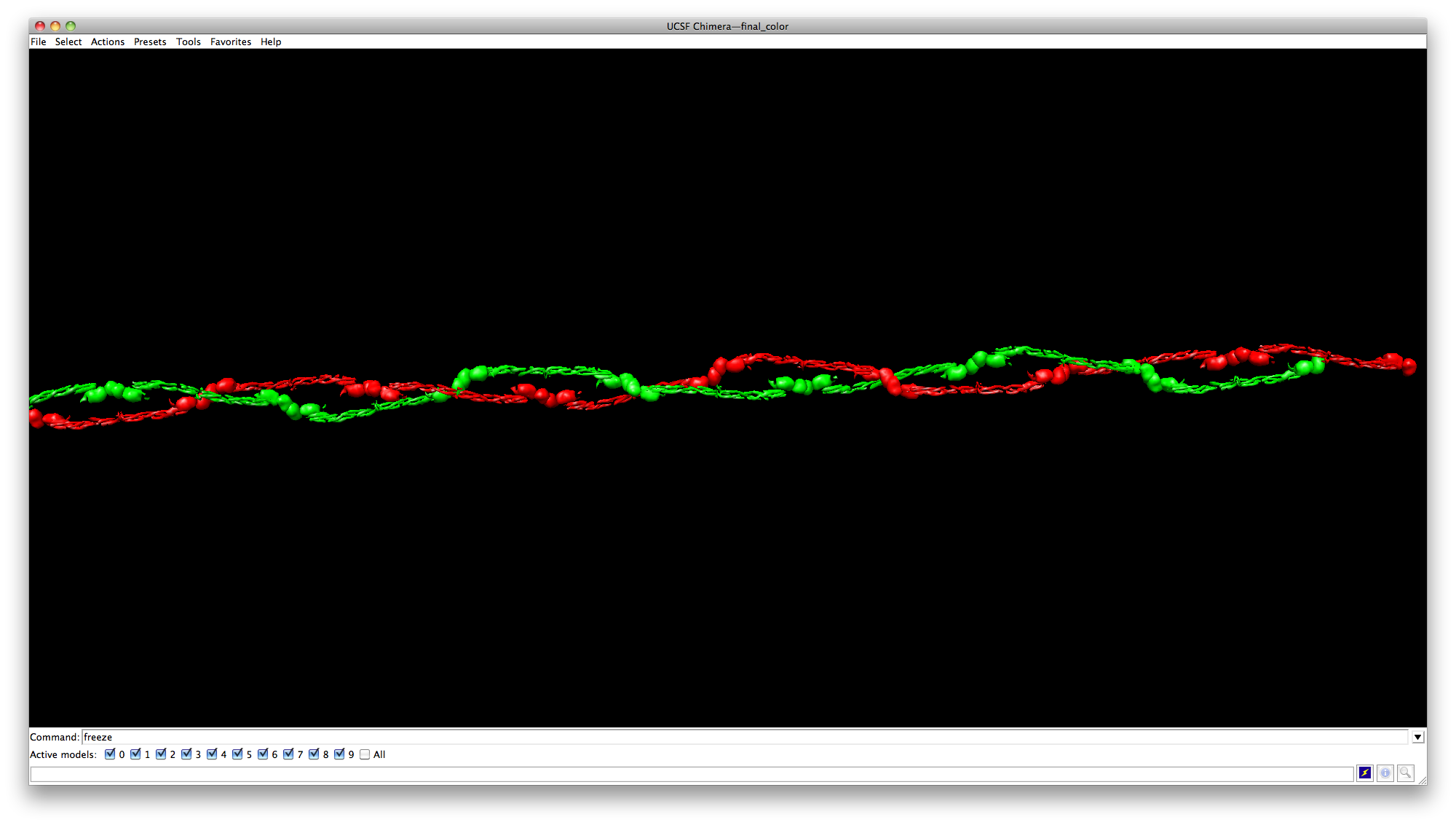
Change colors
Use the same selection trick (Ctrl+shift+mouse click, and then the arrow key “up”) to select one of the chains, like in the image below.
Then click Actions->Actins/Bonds->Color to color the whole chain with red.
Next select the other chain, and color it with green.
Make a movie
To make the object rolling along the X axis, type
roll x
(replace x with three numbers separated by commas for any axis)
To start recording, type
movie record
To stop recording, type
movie stop
To freeze the object, type
freeze
To generate a movie, type
movie encode mformat avi output /whatever path you want/chimela.avi