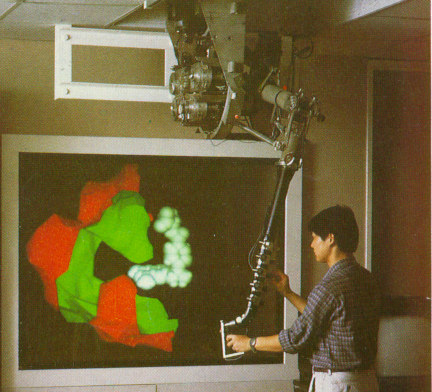
 Ming Ouh-young at UNC designed and built a haptic feedback system to simulate the interaction of a drug molecule with its receptor site in a protein. (Brooks, Ouh-Young et al. 1990; Ouh-young 1990) This system, called the Docker, computed the force and torque between the drug and protein due to electrostatic charges and inter-atomic collisions. These forces were presented to a chemist, pulling the drug towards local energy minima. This task is very similar to that of other “lock and key” applications where a scientist moves one object and senses collisions with other objects in the environment The system presented the force and torque vectors both visually and using haptic feedback. Experiment showed that chemists could perform the rigid-body positioning task required to determine the lowest-energy configuration of the drug up to twice as quickly with haptic feedback turned on compared to using the visual-only representations. (Ouhyoung 1990) Scientists also reported that they felt like they had a better understanding of how the drug fit into the receptor site when they were able to feel the forces.
Ming Ouh-young at UNC designed and built a haptic feedback system to simulate the interaction of a drug molecule with its receptor site in a protein. (Brooks, Ouh-Young et al. 1990; Ouh-young 1990) This system, called the Docker, computed the force and torque between the drug and protein due to electrostatic charges and inter-atomic collisions. These forces were presented to a chemist, pulling the drug towards local energy minima. This task is very similar to that of other “lock and key” applications where a scientist moves one object and senses collisions with other objects in the environment The system presented the force and torque vectors both visually and using haptic feedback. Experiment showed that chemists could perform the rigid-body positioning task required to determine the lowest-energy configuration of the drug up to twice as quickly with haptic feedback turned on compared to using the visual-only representations. (Ouhyoung 1990) Scientists also reported that they felt like they had a better understanding of how the drug fit into the receptor site when they were able to feel the forces.
The Docker application, like other path-planning applications, required the presentation of both force and torque to the user. Because the drug molecule was not a point probe, different portions of it could collide with the protein at the same time. Extricating the drug from a collision sometimes required both translation and twisting. If a chemist were provided with only force (translation) information and no torque (twist) information, they could be led to move the drug in an improper direction.
